Hadoop is pretty famous for analysing huge datasets in batch (MapReduce) or in-memory (Spark). To abstract all the technical things (e.g. setting up a cluster, executing/managing Hadoop programs, HDFS Staging) from end users, Lukas and I developed the Hadoop workflow system Cloudgene, introduced in this blog entry.
Cloudgene + Docker
Even if the installation process of Cloudgene is quite doable, the prerequisite of having a running Hadoop Cluster is kind of a shortcoming. To overcome this, we generated a Hadoop Docker Image to set up a Cloudera Hadoop (CDH5) cluster in pseudo-distributed mode. With the help of this image we are now able to install Cloudgene atop of Hadoop and new applications (apps) can be registered. This means that every random Hadoop program can be connected to Cloudgene and benefit from its workflow capabilities.
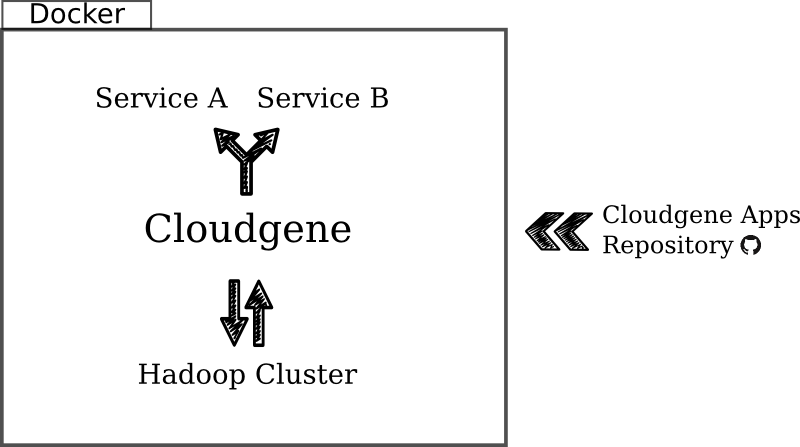
Connecting new apps to Cloudgene
Imagine you are the developer of a new Hadoop application to analyse some kind of (big) data. Since you’re not interested in being the guy who applies your application to all datasets, you want to present your co-workers a “do-it-yourself” service. This is exactly the job of Cloudgene. To register your application within Cloudgene, two things are important:
- Your Hadoop (e.g. MapReduce/Spark) application is executable on the command line (e.g.
hadoop jar wordcount.jar <in> <out>). You can also combine apps written with different technologies (e.g. MapReduce, Spark, Unix Tool, R) to a powerful Cloudgene workflow. - You know the basics of the Cloudgene Workflow Language developed by Lukas to connect your app to Cloudgene. A simple example (again the famous Wordcount) has been introduced in this post.
Pull Cloudgene Image
To deploy your application, simply pull the latest Docker Image of Cloudgene with all dependencies from Docker Hub.
docker pull seppinho/cloudgene-dockerRegister your app
We already prepared an app repository including two sample applications (Wordcount and mtDNA-Server). Cloudgene allows us to register different repositories (containing several apps) making each Cloudgene installation user-specific. Specify your own repository with the --repository command line option.
Run your service
Running this command will (a) configure Hadoop in pseudo-distributed mode, (b) install Cloudgene and all dependencies and (c) register the repository. When accessing the web interface (http://ip-address:8082), applications can now be managed and run graphically. So now it’s the time to simply share the link with your co-workers.
sudo docker run --privileged -it -p 8082:8082 seppinho/cloudgene-docker
--repository https://github.com/seppinho/cloudgene-apps-dockerCloudgene in Real Life
Cloudgene is the underlying framework of the Michigan Imputation Server and is now in production since 1 1/2 years. We imputed over 1.4M Genomes and improved Cloudgene a lot to provide a stable service to the community.
If you are interested to integrate your app into Cloudgene, please let us know.
